Proteins are biologically essential molecules consisting of specific combinations of amino acids. Reverse-phase chromatography is the gold standard for protein analysis, which separates them based on their proportion of hydrophobic amino acids. In this guide, we outline the mechanism of RP-HPLC protein analysis and highlight all 20 amino acids in chromatography.
Various separation techniques are available to analyze peptides and proteins, with the most common being reverse-phase high performance liquid chromatography (RP-HPLC). RP-HPLC separates proteins based on hydrophobic bonding between the column’s stationary phase (hydrocarbon bonded-silica) and the protein’s hydrophobic amino acid residues (leucine, phenylalanine, valine, isoleucine, etc.).
Chromatography for Separating Proteins
What is Protein (Peptide) Analysis?
Proteins (also known as peptides) make us tick. They are involved in all the processes that keep us alive and functioning, which makes them a vital component of biochemical research. Proteins are made up of long strands of amino acids (20 in total) arranged in a specific order. Each amino acid is also known as a ‘residue’.
For example, insulin—a protein that regulates blood sugar levels—comprises 51 amino acid residues. On the other hand, cytochrome P450 3A4 (CYP3A4)—a key enzyme in drug metabolism—is a large protein made up of 503 residues!
Proteins can differ slightly, such as bovine (cow) insulin and human insulin having the same length but different amino acids at three locations. Mutations can also cause such amino acid substitutions. Minor differences in protein length are also possible through insertion/deletion mutations.
Whether for research or quality control purposes, it is crucial to differentiate between proteins. Protein analysis is often done using chromatography, a collection of techniques that separate the proteins based on size, shape, or chemical properties. Once separated, a detector analyzes each protein individually.
Common Protein Chromatography Techniques
When different proteins are mixed in a solution, it is almost impossible to analyze them! Chromatography encompasses various techniques for separating proteins based on specific characteristics. Below are several examples:
- Size-exclusion chromatography separates proteins/peptides based on their overall size. Larger molecular weight proteins elute first, while smaller ones get trapped within the pores of the column.
- Ion-exchange chromatography separates molecules based on their charge. Several acidic and basic amino acid residues are charged when dissolved in water. Proteins with many of these charged residues will bind to the ion-exchange column and elute more slowly.
- Reverse-phase chromatography utilizes the hydrophobic interaction between amino acid residues and non-polar molecules to separate proteins. Proteins with a larger proportion of hydrophobic amino acids elute more slowly.
The gold standard for separating proteins based on their amino acid sequence is reverse-phase HPLC (high-performance liquid chromatography). In this technique, proteins are dissolved in a polar liquid solvent and passed through a reverse-phase column under high pressures.
By separating proteins based on the hydrophobic amino acid residues instead of shape or size, RP-HPLC can differentiate between proteins that have nearly identical structures!
How Reverse-Phase HPLC Works
The Reverse-Phase Column
Most proteins have hydrophilic and hydrophobic regions (see the 20 Amino Acids in Chromatography section below for more information) in their overall structure. Reverse-phase columns exploit these hydrophobic regions by interacting with them through attached hydrophobic (non-polar) molecules, known as the stationary phase.
Reverse-phase columns are made by tightly packing silica (silicon dioxide) particles into a column. Silica alone is not hydrophobic, so long-chain hydrocarbons are attached to the particles. A variety of alkyl chain-bonded silica columns are commercially available:
| Column | Structure | Hydrophobicity |
|---|---|---|
| C18 | Linear 18-carbon chain | Most hydrophobic |
| C8 | Linear 8-carbon chain | Hydrophobic |
| C4 | Linear 4-carbon chain | Least hydrophobic |
Other hydrocarbons can also be attached to the silica, including those containing polar groups that allow for polar interactions with hydrophilic proteins.
The standard column length for RP-HPLC columns is 150 mm. However, column length is not a key factor for separation (resolution) between proteins and peptides, especially for larger proteins with more prominent hydrophobic regions.
For smaller proteins with fewer hydrophobic amino acid residues, a longer column might be needed to increase the contact time between the protein and column surface.
The standard internal diameter for RP-HPLC columns is 4.6 mm, with a default flow rate of 1 ml/min. UHPLC (ultra-high-performance liquid chromatography) systems have column diameters and flow rates of 2.0 mm and 200 μl/min, which means less solvent is required.
Choosing the Mobile Phase Solvent
A critical factor in RP-HPLC is the choice of solvent. For successful chromatography analysis of proteins, all of the components need to dissolve in whatever liquid is chosen as the mobile phase. Since the hydrophobic residues interact with the stationary phase, the solvent should be hydrophilic (polar) to counteract this.
For more information on choosing the right mobile phase solvent, read our guide on solubility and dielectric constants.
We can use a gradually changing mixture of solvents throughout the chromatography run, also known as a solvent gradient. Solvent gradients generally increase in polarity over time, such that the least hydrophobic proteins/peptides elute first, while the most hydrophobic ones are only released near the end of the run by the polar mobile phase.
Below is a table of several chemicals used in mobile phase solvent systems, from least to most polar, along with a short description.
| Chemical | Utility as RP-HPLC Mobile Phase |
|---|---|
| Tetrahydrofuran | Used with aqueous mixtures to dissolve a wide range of proteins |
| Isopropanol | Useful for dissolving very hydrophobic proteins |
| Acetone | Used as an additive, but absorbs in the range of many amino acid residues (if detected by UV) |
| Methanol/Ethanol | Rarely used for protein/peptide analysis (but cost-effective) |
| Acetonitrile | Favored mobile phase solvent, can dissolve most proteins with good RP-HPLC performance |
| Water | Most polar but should not be used alone in RP-HPLC as it can desorb the hydrocarbon chains in the column |
Another critical factor for RP-HPLC solvents is the need for an ion-pair reagent to improve the resolution between proteins. These reagents dissociate into positively and negatively charged species in solution, ‘pairing’ themselves with charged amino acid residues in the mobile phase. This stabilizes them and makes them less hydrophilic, which improves their bonding with the stationary phase.
Only a low concentration (less than 0.1% v/v) is enough for ion-pair reagents to achieve their stabilizing effect. Common ion-pair reagents for basic samples:
- Tetraalkylammonium hydroxides/bromides/chlorides/sulfates/phosphates like tetraethylammonium hydroxide, tetrabutylammonium phosphate, etc.
- Dialkylammonium acetates
Common ion-pair reagents for acidic samples:
- Tri-/penta-/hepta-/nona-fluoroacetic acid (TFA is by far the most commonly used)
- Sodium alkyl sulfonates
- Sodium dodecyl sulfate
Detectors for Protein Analysis
UV detectors are the most widely used for protein analysis, as peptide bonds absorb in this range (215 nm). Some amino acid residues also contain UV-active groups (tyrosine, tryptophan and phenylalanine have aromatic phenol groups) that absorb strongly in this region, thanks to their conjugated pi-electron structures.
However, detection by UV absorption can be an issue depending on the solvent system used for the mobile phase. Certain chemicals like acetone and trifluoroacetic acid will absorb in the UV region, which can cause poor resolution in the chromatogram and baseline drift (if used in a solvent gradient).
Mass spectrometers can detect molecule fragments based on their charge/size, making them versatile. Recent advancements in MS technology (matrix-assisted, electrospray ionization) have improved the sensitivities of mass spectrometers, enabling them to distinguish between individual amino acids in a protein.
Structure and Properties of the 20 Amino Acids in Chromatography
The table below shows the chemical structure and hydrophobicity value for all 20 naturally occurring amino acids. The hydrophobicity and abundance values are relative units and should be used as a reference for comparing amino acids in chromatography setups for protein analysis.
| Amino Acid (3-letter/1-letter code) | Chemical Structure | Abundance (1 = least common) (20 = most common) | Hydrophobicity (1 = least hydrophobic) (20 = most hydrophobic) | Unique Characteristic |
|---|---|---|---|---|
| Alanine (Ala/A) |  | 19 | 14 | Alkyl residue |
| Arginine (Arg/R) | 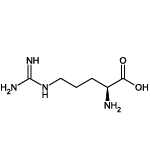 | 10 | 1 | Positively charged |
| Asparagine (Asn/N) | 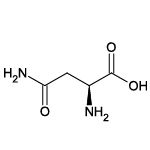 | 8 | 7 | Contains -NH2 group |
| Aspartic acid (Asp/D) | 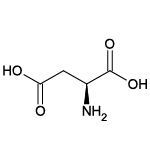 | 11 | 3 | Negatively charged |
| Cysteine (Cys/C) | 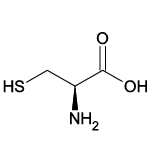 | 2 | 12 | Contains sulfur atom |
| Glutamic acid (Glu/E) |  | 15 | 5 | Negatively charged |
| Glutamine (Gln/Q) | 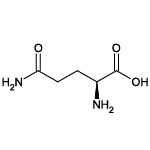 | 6 | 4 | Contains -NH2 group |
| Glycine (Gly/G) | 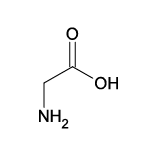 | 17 | 13 | Smallest, only -H residue |
| Histidine (His/H) | 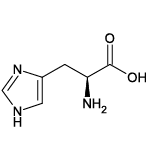 | 3 | 7 | Positively charged |
| Isoleucine (Ile/I) | 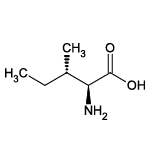 | 13 | 20 | Alkyl residue |
| Leucine (Leu/L) | 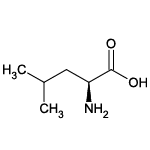 | 20 | 17 | Alkyl residue |
| Lysine (Lys/K) | 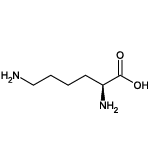 | 14 | 2 | Positively charged |
| Methionine (Met/M) | 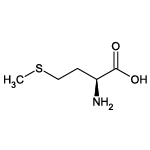 | 4 | 15 | Contains sulfur atom |
| Phenylalanine (Phe/F) |  | 7 | 19 | UV-active |
| Proline (Pro/P) | 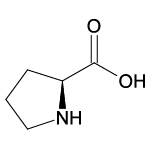 | 9 | 10 | Contains cyclic -NH2 group |
| Serine (Ser/S) | 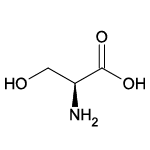 | 18 | 8 | Contains -OH group |
| Threonine (Thr/T) | 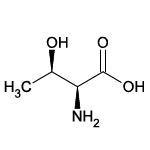 | 12 | 9 | Contains -OH group |
| Tryptophan (Trp/W) | 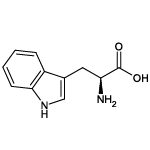 | 1 | 16 | UV-active |
| Tyrosine (Tyr/Y) | 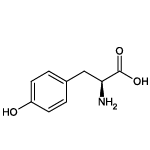 | 5 | 11 | Contains -OH group, UV-active |
| Valine (Val/V) | 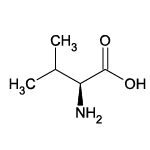 | 16 | 18 | Alkyl residue |
About the Author

Sean is a consultant for clients in the pharmaceutical industry and is an associate lecturer at La Trobe University, where unfortunate undergrads are subject to his ramblings on chemistry and pharmacology.